Data Searches
To access the different data searches, click on the Search menu in the header and then select the data type you would like to search (Fig. 7). To learn more about each search interface, please see the Outline link below the figure.

Figure 7. Search menu in header provides links to different searches.
Genes and Transcripts
The Genes and Transcripts Search is located under the Search menu in the header. The Genes and Transcripts Search allows you to search sequences that are available in CGD with several different parameters. A few parameters can be used to return a broad range of results, or numerous parameters can be used to find very specific data. Searches can be limited to a certain genus and species. Once a genus has been selected, the species list will be populated and then a species selection can be made (Fig. 8A). Searches can also be limited to datasets, such as genome and RefTrans assemblies or NCBI genes. For a genome assembly dataset, the search can be restricted to a chromosome or scaffold and further restricted to a region on that chromosome or scaffold (Fig. 8B). Searches for a specific gene or transcript name, or a list of names that are uploaded as a text file, are also possible with this search interface.

Figure 8. CGD Genes and Transcript Search interface.
All search results are returned as a table with hyperlinks to more info (Fig. 9). The table can be downloaded and a Fasta file of the returned sequences can also be downloaded (Fig. 9A). To do another search, click reset.

Figure 9. Gene and Transcript Search results table
Germplasm
The Germplasm Search can be accessed through the Search menu in the header. Information about germplasm linked to data in CGD can be searched by species (Fig. 10A) and/or name (Fig. 10B). A list of germplasm names, in a text file, can also be uploaded to search for multiple germplasm at once.

Figure 10. CGD Germplasm Search interface.
Results are returned in a table format that can be downloaded (Fig. 11A). Blue text indicates hyperlinks to more detailed information (Fig. 11B). Searches can be refined by editing the parameters or a new search can be initiated by clicking the reset button.

Figure 11. Germplasm Search results
Markers
To search the markers in CGD, click on the Search menu in the header and select Marker Search. The Marker Search is useful for retreiving information about markers and can be a broad search or a very specific search depending on the number of parameters used. Markers can be searched by typing in a marker name, or a text file of multiple marker names can be uploaded to search for multiple markers at once (Fig. 12). The search can also be restricted by the marker type (Fig. 12A), the species the marker was developed in, or the species the marker was mapped in. To find out more information about the marker types, click the question mark logo next to Marker Type. For markers that have been mapped to a genetic map, the search can be restricted to a certain linkage group (Fig. 12B). And the search can be even further restricted to a certain locations on the linkage group.

Figure 12. CGD Marker Search interface
The search results are returned in a table format (Fig. 13). There are hyperlinks within the table that take you to more details about the marker or where the marker is mapped. This data table can be downloaded in a format that can be easily opened in applications such as Excel (Fig. 13A). Clicking on the marker name (Fig. 13B), displays the marker page which has information such as alignments and map positions. If you would like to change the search, either edit the parameters or click the reset button to start all over.
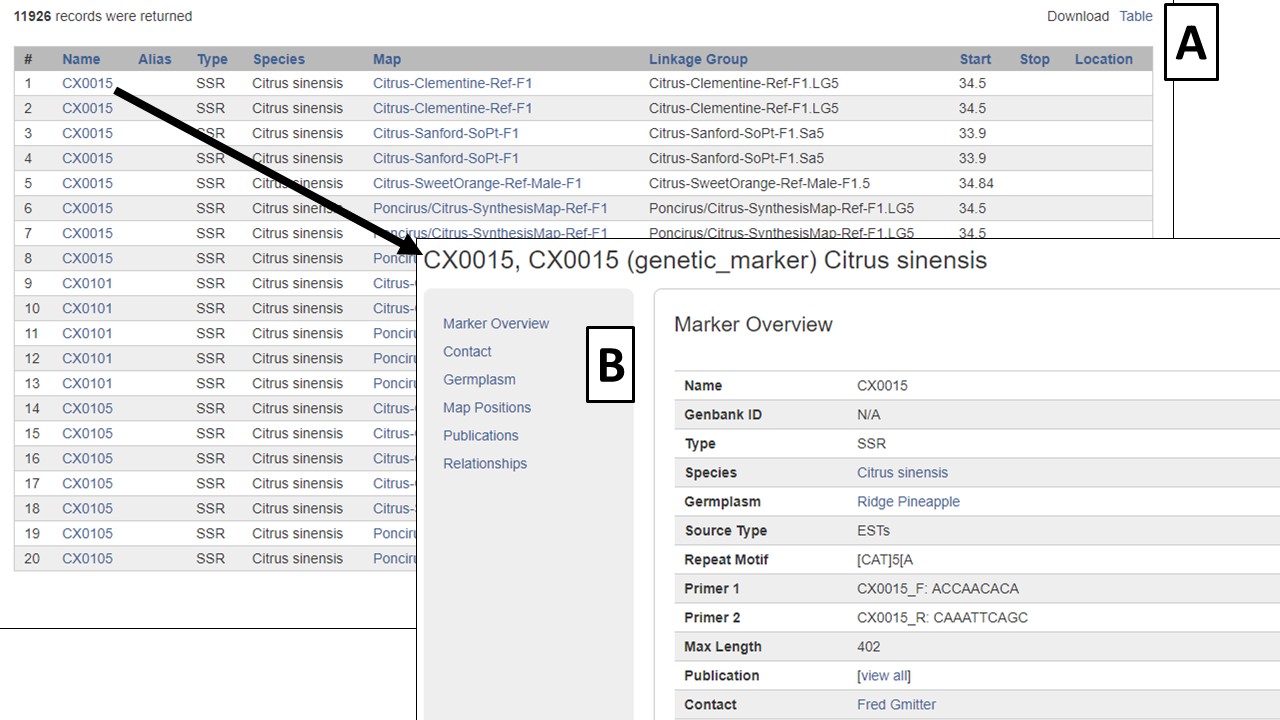
Figure 13. Marker Search results
Publications
To search publications on CGD, click on the Search menu and select Publication Search. The Publication Search is like many common literature searches, but is limited to publications that have been added to CGD. The publications can be searched by using keywords within different fields (Fig. 14A). Multiple fields and keywords can be entered and more fields can be added by clicking the Add/Remove buttons (Fig. 14B). A range of years can also be entered (Fig. 14C).
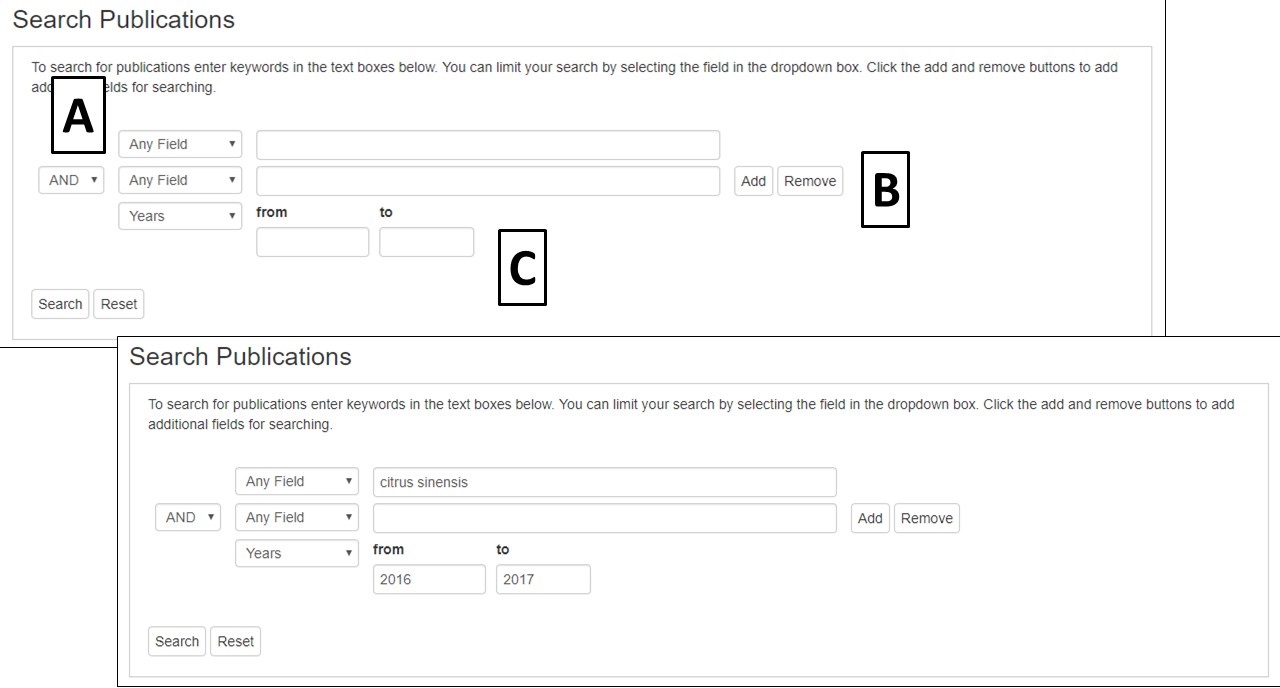
Figure 14. CGD Publication Search interface
The search results table has information about each publication (Fig. 15A). By clicking the publication title, more detailed information is displayed. Most publications also have a link to the publisher website or PubMed record in their titles (Fig. 15B).

Figure 15. Publication search results and detailed publication information
QTLs and MTLs
Quantitative trait loci (QTLs) and Mendelian trait loci (MTLs) that are entered in CGD are searchable from the QTL Search option under the Search menu in the header. The search can be restricted by QTL or MTL as well as by species (Fig. 16A). A certain trait ontology category can be selected to limit the returned results to only that trait type. If you are looking for a specific trait, you can do a keyword search of the trait names or search by the published name or the CGD assigned name (Fig. 16B). Please see the CGD Trait Abbreviation Table for more information about the trait abbreviations used in CGD.

Figure 16. CGD QTL Search interface
The results are returned in a table that has blue hyperlinks to more information about the QTL or MTL, the map it is located on, and the species it is from (Fig. 17). The table can also be downloaded (Fig. 17A). Clicking on the QTL label, opens the QTL information page which has a link to map positions (Fig. 17B). From the map position table, the QTL can be viewed in MapViewer or CMap.

Figure 17. CGD QTL Search results
Sequences
To search sequences, click on the Search menu on the homepage header and select Sequence Search. This search is useful for retrieving information about certain sequences from a larger dataset, or all the sequences from one or more datasets. The Gene and Transcript Search page is for searching genes and transcripts only, whereas the Sequence Search page also includes other sequence types. A few search parameters can be used to return a broad range of results, or more parameters can be selected to find very specific data. This is not a BLAST search, BLAST is available under the Tools menu.
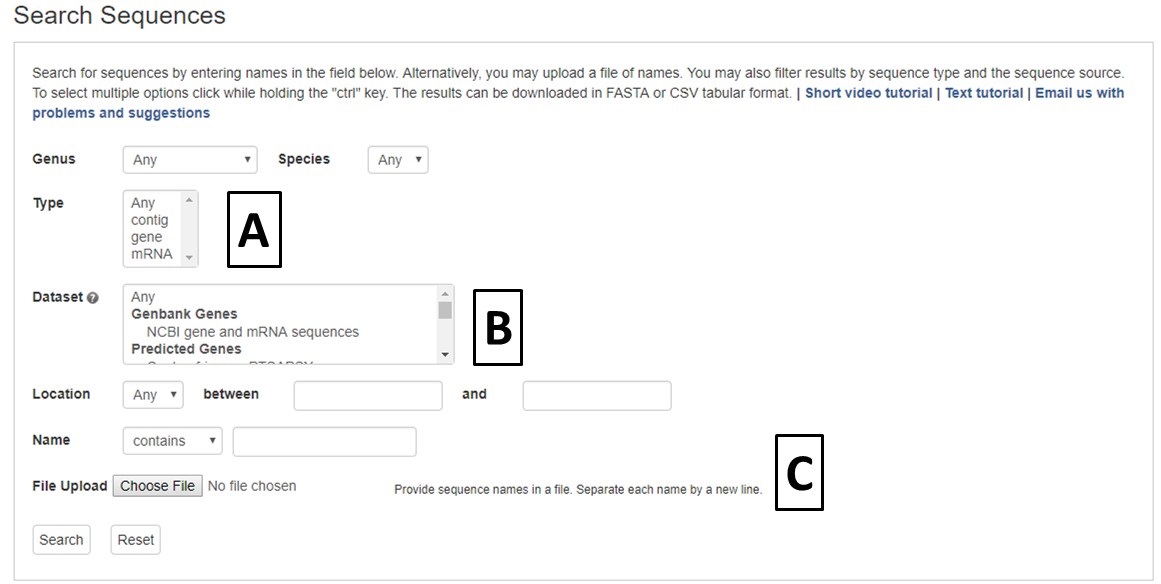
Figure 18. CGD Sequence Search interface
Sequence searches can be limited by genus and species, or sequence type (Fig. 18A). The search can also be restricted to a certain genome dataset and further restricted to a certain chromosome or scaffold location for within that assembly (Fig. 18B). The sequences in CGD can also be searched by sequence name, and there is an option to upload a text file of sequence names (Fig. 18C).

Figure 19. Search results table
All search results are returned in a table with hyperlinks to more information (Fig. 19A). The results table can be downloaded and a Fasta file of the sequences on the table can also be downloaded (Fig. 19B). To do a different search, either edit the parameters and search again, or click the Reset button.