Tools
To access the different Tools available on CGD, click on the Tools menu in the header and then select the tool you want to use (Fig. 20A). Many of the tools are also quickly accessed through links in the Tools Quick Start (Fig. 20B). To learn more about each search tool, please see the links below the figure.

Figure 20. Tools menu in header and Tools Quick Start on homepage
BLAST
CGD offers BLAST with sequence databases from sweet orange, clementine, mandarin, and trifoliate orange. Genome, unigene, and reference transcriptome assemblies for the citrus species are available along with complete genomes of Ca. Liberibacter and Liberibacter crescens. The BLAST interface looks and functions like the interface available on NCBI and information on the different settings and how to use BLAST can be found in the BLAST Help manual. The new Tripal BLAST module displays the results in a interactive interface (Fig. 21A) and for alignments to the genome scaffolds that are also in the CGD JBrowse, there is a link to view the BLAST hit in JBrowse (Fig. 21B). Features that are in the CGDdatabase (CDS, peptides, unigenes, RefTrans), will have links to more information in CGD.
.jpg)
Figure 21. BLAST results on CGD
JBrowse
CGD has an instance of the JBrowse genome browser for viewing genome data. A list of the genomes available in CGD can be accessed by clicking the JBrowse link in the Tools menu. Please watch the JBrowse tutorial for more details about how to navigate and use JBrowse.
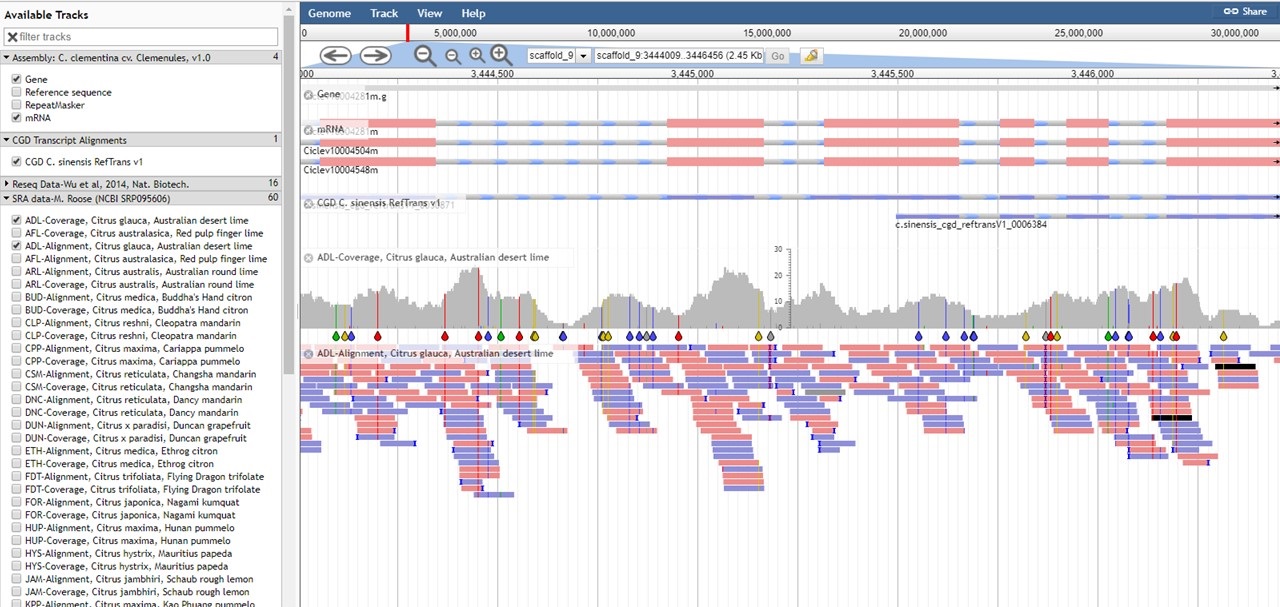
Figure 22. JBrowse of C. clementina genome.
For the tracks of aligned reads (BAM files) in the C. clementina v1.0 genome, the table below describes what the read colors mean in JBrowse.
| Aligned Read Color Meaning | Color of Read |
|---|---|
| Forward Strand | |
| Reverse Strand | |
| Forward strand missing mate | |
| Reverse strand missing mate | |
| Forward strand not proper | |
| Reverse strand not proper | |
| Forward strand on different chromosome | |
| Reverse strand on different chromosome |
CitrusCyc
CitrusCyc pathways were generated using Pathway Tools and are available under the CitrusCyc link in the Tools menu. Pathway Tools allows users to view metabolic pathways that are in genomes. Please see the manual for Pathway Tools for more information on use.

Figure 24. Citrus clementina cellular overview in CitrusCyc on CGD
MapViewer
MapViewer is a new tool for viewing genetic maps on CGD. It can be accessed from the Tools menu in the header (Fig. 25A), the Species Overview page (Fig. 25B), or the Map Overview page. The Map Overview page displays a summary graphic of all linkage groups (Fig. 25C) and clicking a linkage group opens a more detailed view in MapViewer.

Figure 25. Different ways to open MapViewer.
MapViewer displays the complete linkage group on the left, and the selected region on the right (Fig. 26). The selected region can be changed by dragging and resizing a window on the complete linkage group on the left side. There is a legend of the marker colors below the linkage group figure (Fig. 26A). Information about the markers is displayed in the upper right corner when the pointer is over a marker name on the right side graph. Clicking on the marker name on the rights side graph, opens the marker details page.

Figure 26. MapViewer displays a static linkage group graph on the left and a dynamic graph on the right.
A different map or linkage group can be displayed using the controls at the bottom of the MapViewer page (Fig. 27A). The color of the markers and which markers are displayed can be changed with the controls (Fig. 27B). The ruler and marker positions can also be toggled on or off (Fig. 27C). After changing any of the four parameter sections, the Submit button must be pressed to display the changes.
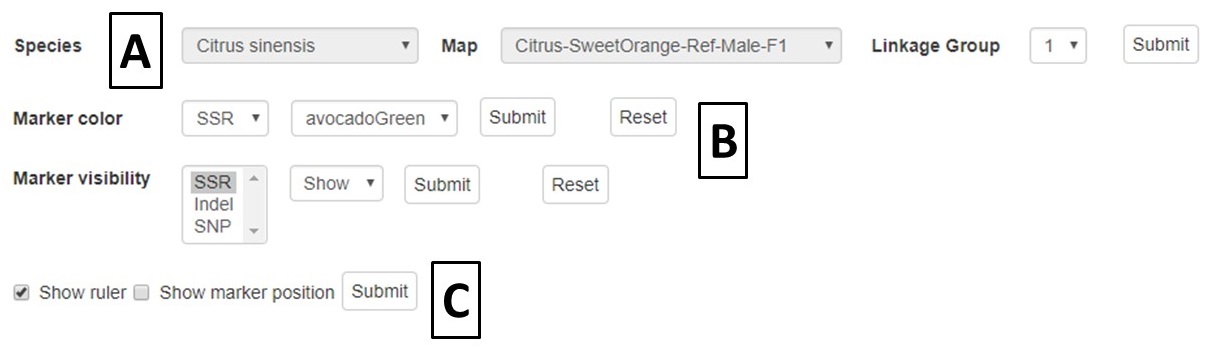
Figure 27. MapViewer control panel.
Pictures from MapViewer
MapViewer allows users to export figures as a high-resolution PNG file that is suitable for publication. Items that can exported have a clickable camera icon that will trigger a file download. Users can export the map overview which has all the linkage groups (Fig. 1), a single linkage group (Fig. 2), a linkage group comparison (Fig. 3), a dot plot (Fig. 4) or a correspondence matrix (Fig. 5).

Figure 1. How to download a map overview.

Figure 2. How to download a linkage group figure.
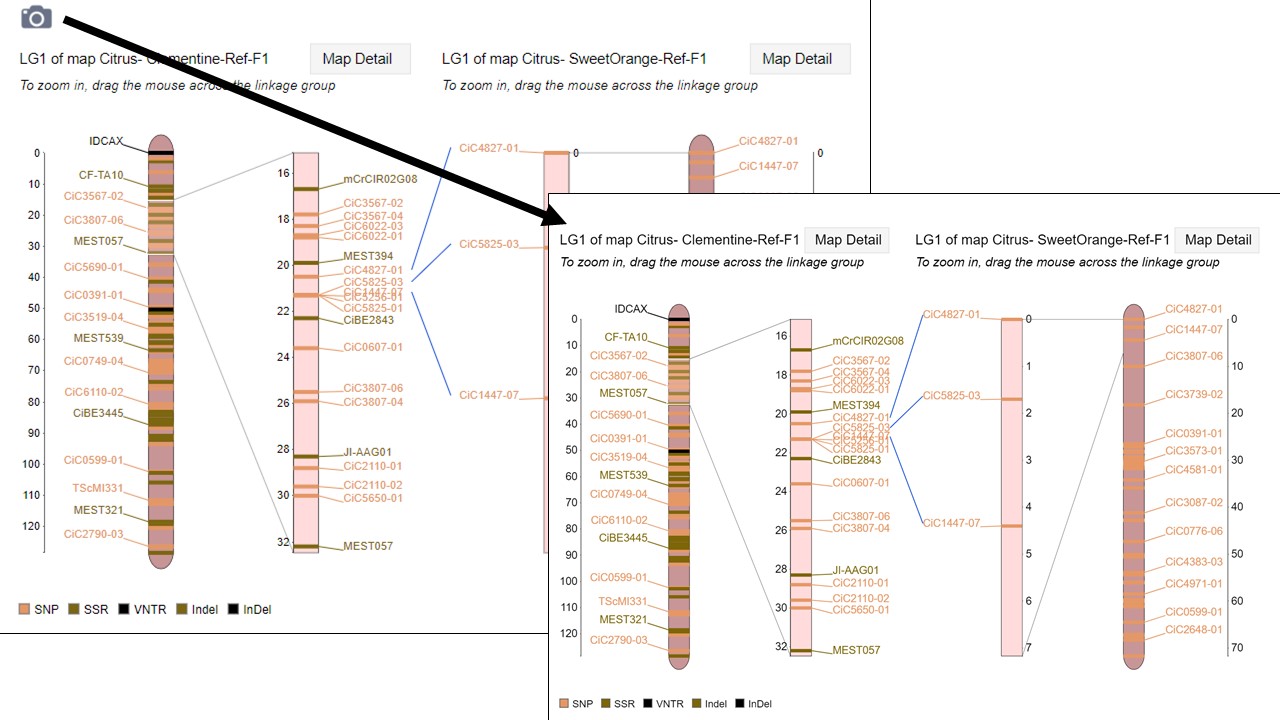
Figure 3. How to download a linkage group comparison figure.

Figure 4. How to download a linkage group comparison dot plot.

Figure 5. How to download a linkage group comparison correspondence matrix.
Synteny Viewer
CGD uses the Tripal Synteny Viewer, developed by the Fei Bioinformatics Lab, to display the analysis results of Citrus and Liberibacter genomes that were compared using the program MCScanX. Synteny Viewer is accessed under the "Tools" menu (Fig. 1A) or via a link on the Species Overview page.
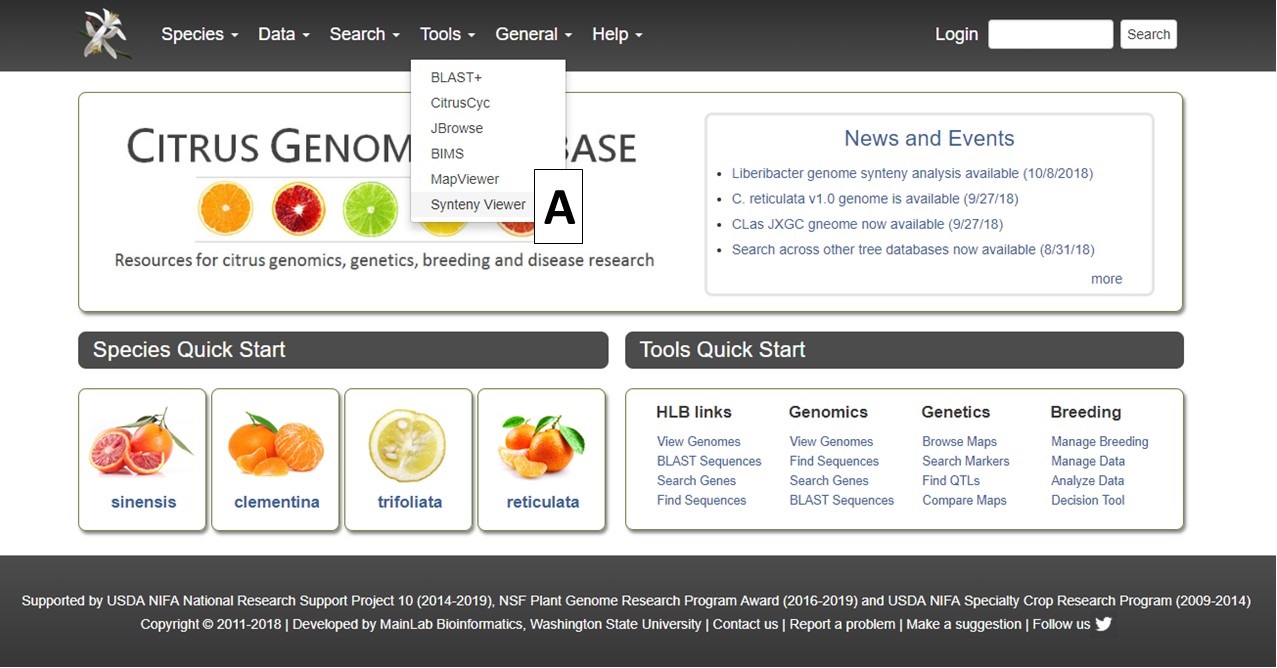
Figure 1. Accessing Synteny Viewer on CGD.
On the Synteny Viewer interface, there is some information about the tool and a pull-down menu to select the Organism Type (Fig. 2A) of "Plant" or "Bacteria" (Fig. 2B). The "Plant" option allows for the comparison of Citrus sp. genomes and the "Bacteria" option is for comparing genomes of the Ca. Liberibacter sp. and Liberibacter sp. If the block ID number is already known, the block ID value can be input directly to return just those results.

Figure 2. Selecting plant or bacteria genome option.
Once the organism is selected, another pull-down menu becomes available to select the first genome for comparison (Fig. 3A). After selecting the genome, the "Chromosome/Scaffold" menu will populate with the names of the appropriate scaffolds or chromosomes and one of the sequences can be selected (Fig. 3B). The final option is to select one or more genomes for comparison (Fig. 3C), and then the "Search button" is clicked to start the search (Fig. 3D).
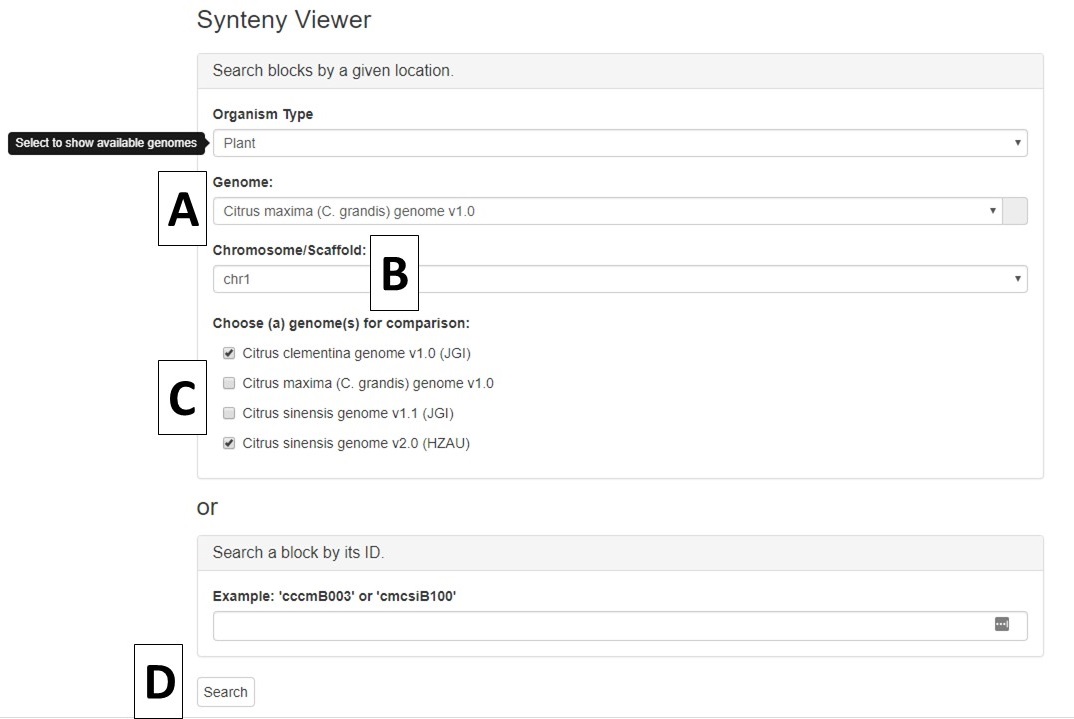
Figure 3. Options for Synteny Viewer search.
When the search is complete, a new page opens with the results. There is a summary of the input settings at the top of the page (Fig. 4A). When multiple genomes are queried, there are tabs to switch between the results (Fig. 4B) and the circular graph (Fig. 4C) will change when a different genome is selected. The syntenic regions are indicated on the circular graph by gray lines. When the mouse hovers over the gray line, a summary is displayed (Fig. 4D). Clicking on the gray lines, opens a page with more details (see below, Fig. 6).
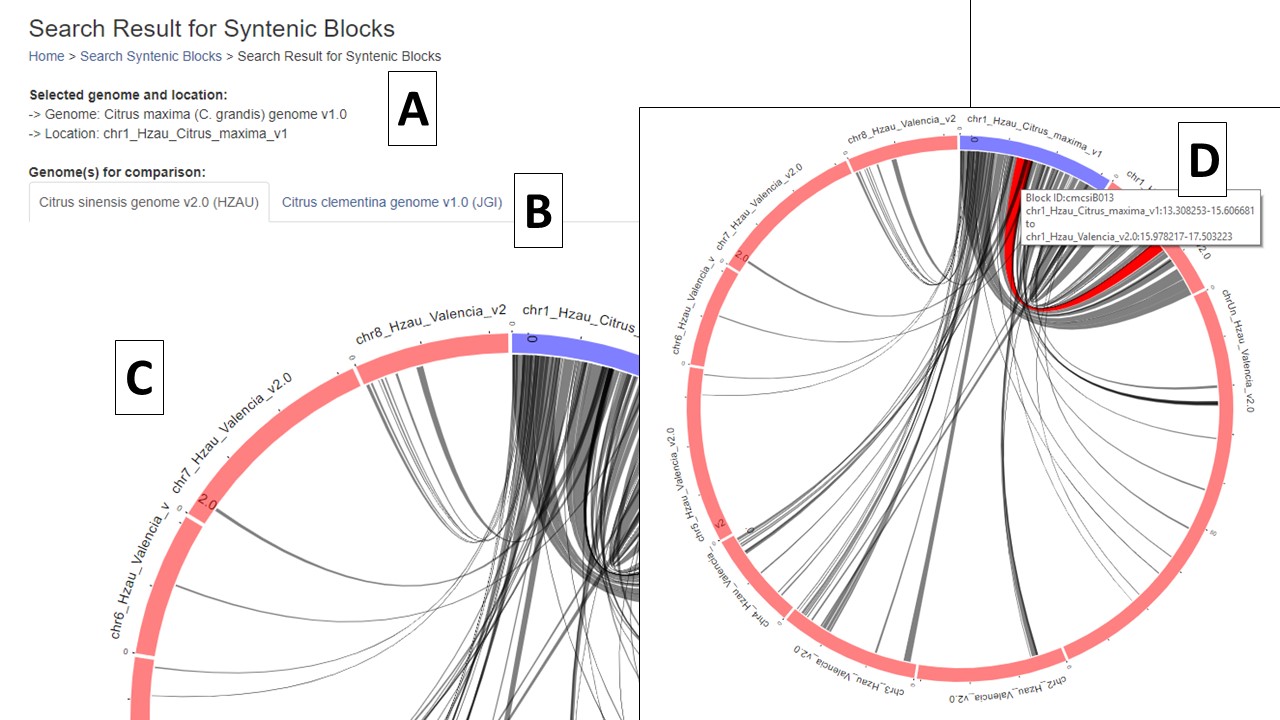
Figure 4. Synteny results and circular graphs.
Under the circular graph is a table listing the syntenic blocks that are displayed as gray lines in the circular graph. Clicking on the block name (Fig. 5A) opens the same details page as clicking on the gray lines on the circular graph.
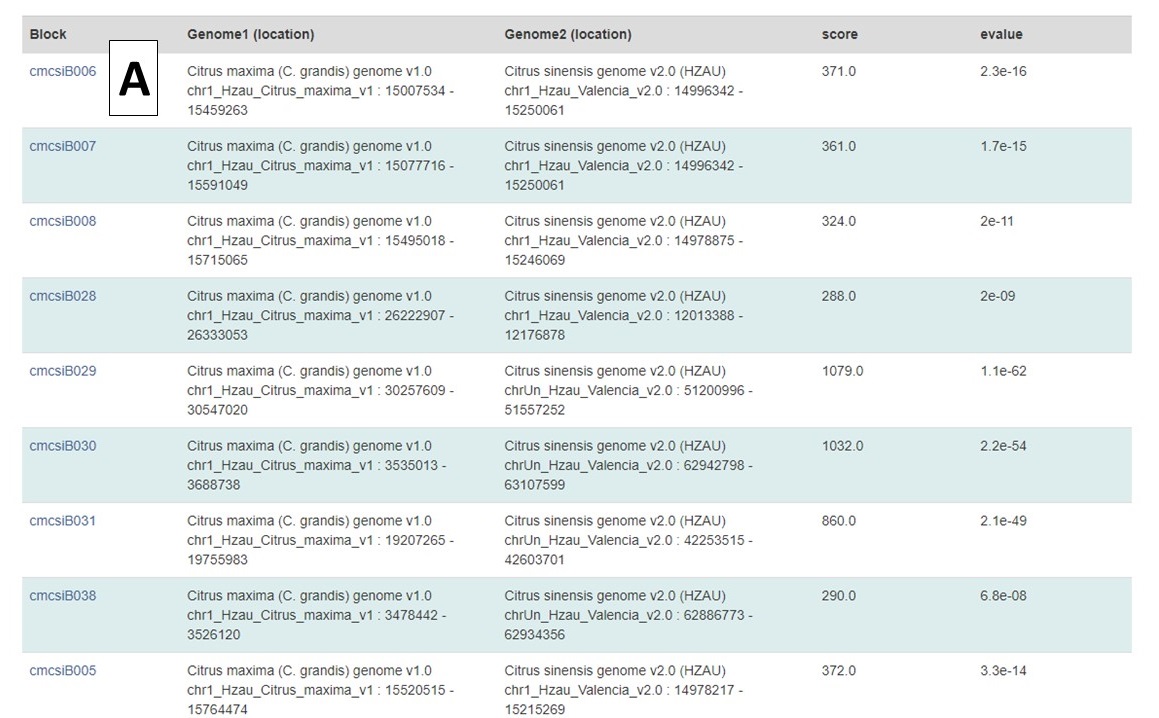
Figure 5. Synteny block list.
The syntenic block details page has an overview section at the top listing the details for each genome in the comparison (Fig. 6A), a side-by-side graphic of the syntenic block from each genome (Fig. 6B), and a table showing the genes in the syntenic block from each genome (Fig. 6C). Clicking on the gene name in either the table or on the graphic will open the feature details page from the CGD database.
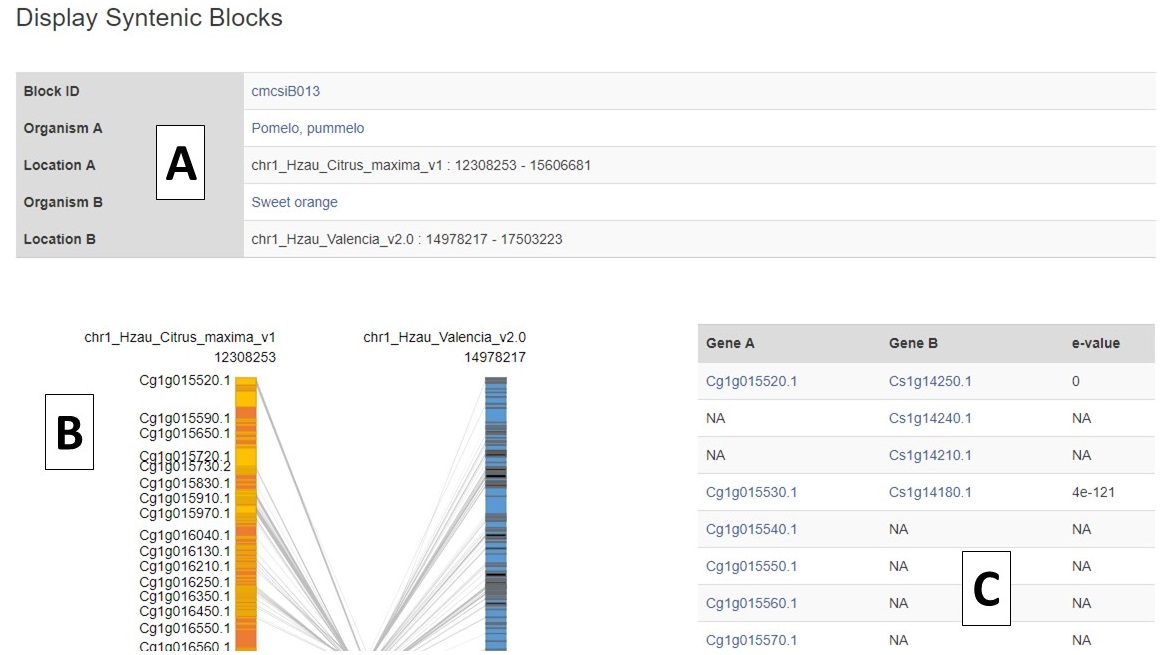
Figure 6. Syntenic Block Details Page.
The side-by-side graph can be zoomed (Fig. 7) using the scroll wheel on the mouse and the view can be shifted up or down by click and dragging.
.jpg)
Figure 7. Zoomed in view of graph.