User Manual
This manual describes how to access data and use the tools on the Citrus Genome Database (CGD). Please use the sidebar on the left to navigate to different parts of the manual, or click on the "Outline" menu in the lower right. You can access the next page of the manual by clicking on the title of the next page below.
Homepage Overview
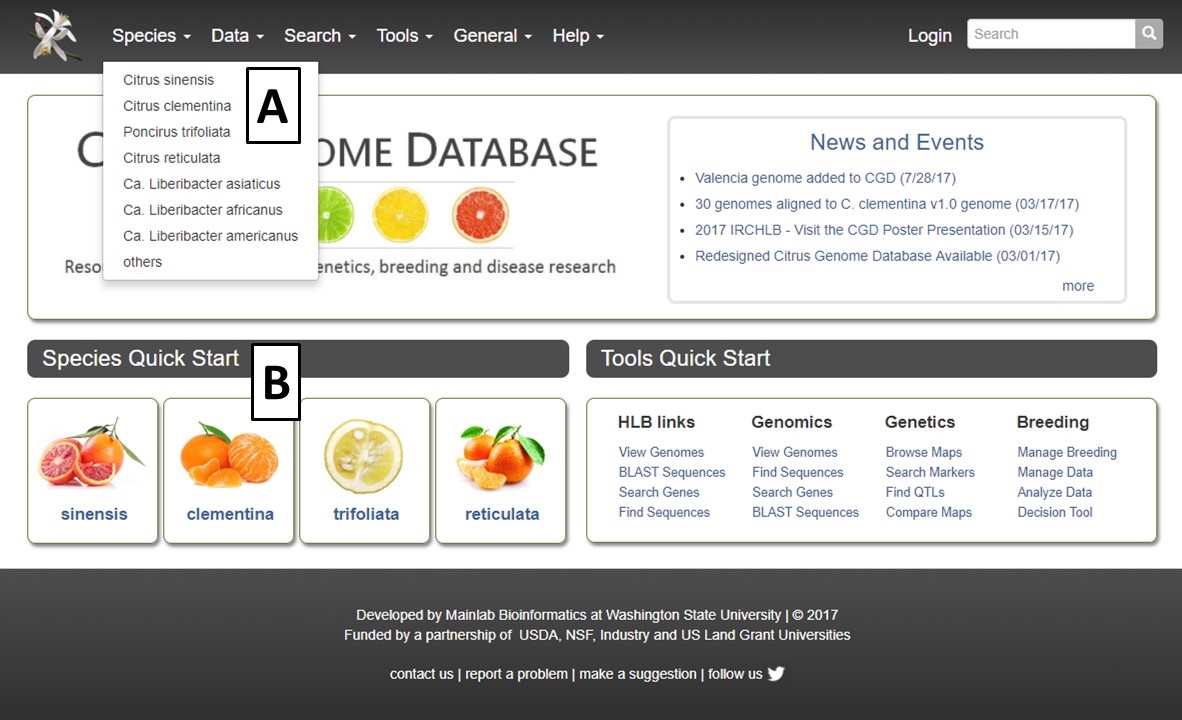
The Citrus Genome Database (CGD) homepage is divided into a few key areas to help facilitate navigation. The Species Quick Start (Fig. 1A), has icons for each species represented in CGD. Clicking on the icon takes you to the Species Overview page (see Species Overview tutorial) which allows for quick access to all the data associated with that crop. To the right of the Species Quick Start is the Tools Quick Start (Fig. 1B). This area has links to common tools and data search interfaces. The center region of the homepage contains a News and Events section (Fig. 1C) which has news about CGD as well as from the community. The homepage also has the traditional pull-down menu bar in the header (Fig. 1D) which also provide access to the species overview pages, data, search pages, tools, and general information.
.jpg)
Figure 1. CGD Homepage
The items under the Search and Tools menus are discussed in other tutorials, but we want to highlight the Data and General menus in this section of the tutorial. Under the Data menu (Fig 2A), you can see a data overview, learn how to submit data, download data, view publication datasets and view information on CGD trait and marker type abbreviations. A summary overview of all data on CGD which can be broken down by species and/or data type is found on the Overview page. Under Data Submission, there are details about submitting data to CGD. CGD will accept published data and we highly recommend contacting us before starting to fill in the data templates. There are two links to the contact form on this page.
The Data Download page contains links to all the various data types in CGD. The links redirect you to either a search page, analysis page, or a table where the information about the data and links to download files are located. For the RefTrans, Unigene, and genome data, the links take you to the analysis page with details about the data and these pages have a link on the left for the download page. For germplasm, markers, QTLs, transcripts and sequences, the links take you to the search page for that data. On the search page, choose your parameters and click submit to retrieve data. The resulting table can then be downloaded.
The Trait Abbreviations page contains the QTL traits that are in CGD and the abbreviations that are used for the internal CGD database generated QTL ID. We have tried to keep our abbreviations consistent with those used in the trait and crop ontologies. On the Marker Types page, there is a table of the marker types that are in CGD. The markers have been generally grouped by the same technologies and we have provided a link to a reference that describes the marker technology.
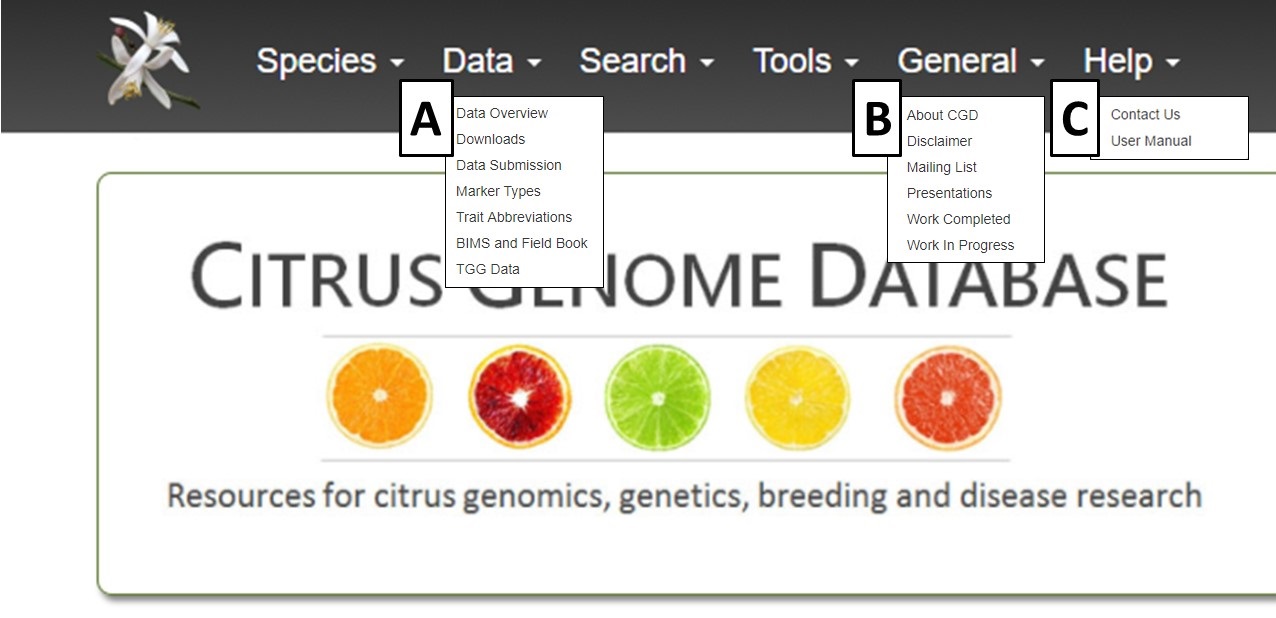
Figure 2. Data, General, and Help menu items.
The General menu (Fig. 2B) contains links to a variety of different items. If you would like to subscribe to the CGD mailing list, please click on Mailing Lists and complete the web form. The General menu also has links to past presentations about CGD, work that is in progress and completed, and how to reference CGD. The Help menu (Fig. 2C) has a link to the CGD User Manual and video tutorials. There is also a Contact Us link that opens a fillable web form. We appreciate input on the website as well as reports of website bugs and data errors.
Species Overview Page
All the available data for each species is easily found through the Species Overview Page. From the CGD homepage, the species overview pages are accessed either by clicking on the Species menu in the toolbar and selecting the species of interest (Fig. 3A) or clicking on the species under the Species Quick Start section (Fig. 3B).
Figure 3. Accessing the Species Overview Pages from the homepage.
On the Species Overview Page, there are two main sections. The left side toolbar is static and has a Data section and a Tools section (Fig. 4A). Clicking on the links in these sections either changes the information to the right of the toolbar (Fig. 4B), or opens another tab with the linked information. The Species Overview Page defaults to the Overview section for each species which contains basic information and a summary of the available data in CGD.

Figure 4. CGD Species Overview Page
Many of the left toolbar links will dynamically change the content to the right of the toolbar (Fig. 5). The Genomes link displays links for the available genomes. On the Genomes pane, the genome name is linked to the analysis page which has detailed information and citations. The second link opens JBrowse to visualize the data. For Germplasm, clicking the link will display a list of germplasm in CGD. Blue text indicates a hyperlink that will display more detailed information. For Transcripts, a table with summary information for the RefTrans and Unigene assemblies is displayed along with links to more detailed information. Under the tools section, the CMap link displays a table that has a list of maps for that species and the reference. The map name has a hyperlink directly to the CMap drawing for that map.

Figure 5. Dynamic data
The rest of the left toolbar links open new tabs. As an example, the Genetic Maps link opens a new tab that has a table of all the genetic maps for the species (Fig. 6). Blue text on the table indicates links to detailed information. The Markers, Publications, Sequences, and Trait Loci links open search interfaces for the selected data type. Please see the separate tutorials for more information on how to use the different data searches and tools.
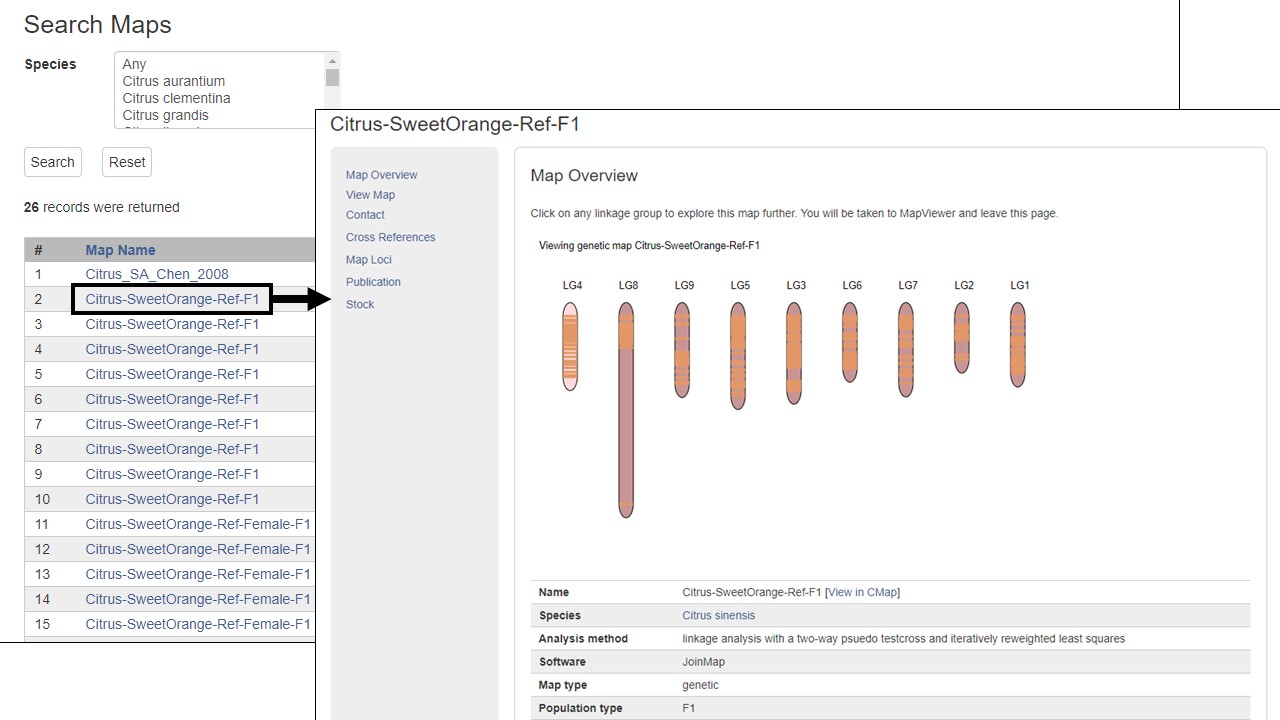
Figure 6. Genetic Map page and example of detailed information available under hyperlink indicated by blue text.
Data Searches
To access the different data searches, click on the Search menu in the header and then select the data type you would like to search (Fig. 7). To learn more about each search interface, please see the Outline link below the figure.

Figure 7. Search menu in header provides links to different searches.
Genes and Transcripts
The Genes and Transcripts Search is located under the Search menu in the header. The Genes and Transcripts Search allows you to search sequences that are available in CGD with several different parameters. A few parameters can be used to return a broad range of results, or numerous parameters can be used to find very specific data. Searches can be limited to a certain genus and species. Once a genus has been selected, the species list will be populated and then a species selection can be made (Fig. 8A). Searches can also be limited to datasets, such as genome and RefTrans assemblies or NCBI genes. For a genome assembly dataset, the search can be restricted to a chromosome or scaffold and further restricted to a region on that chromosome or scaffold (Fig. 8B). Searches for a specific gene or transcript name, or a list of names that are uploaded as a text file, are also possible with this search interface.

Figure 8. CGD Genes and Transcript Search interface.
All search results are returned as a table with hyperlinks to more info (Fig. 9). The table can be downloaded and a Fasta file of the returned sequences can also be downloaded (Fig. 9A). To do another search, click reset.

Figure 9. Gene and Transcript Search results table
Germplasm
The Germplasm Search can be accessed through the Search menu in the header. Information about germplasm linked to data in CGD can be searched by species (Fig. 10A) and/or name (Fig. 10B). A list of germplasm names, in a text file, can also be uploaded to search for multiple germplasm at once.

Figure 10. CGD Germplasm Search interface.
Results are returned in a table format that can be downloaded (Fig. 11A). Blue text indicates hyperlinks to more detailed information (Fig. 11B). Searches can be refined by editing the parameters or a new search can be initiated by clicking the reset button.

Figure 11. Germplasm Search results
Markers
To search the markers in CGD, click on the Search menu in the header and select Marker Search. The Marker Search is useful for retreiving information about markers and can be a broad search or a very specific search depending on the number of parameters used. Markers can be searched by typing in a marker name, or a text file of multiple marker names can be uploaded to search for multiple markers at once (Fig. 12). The search can also be restricted by the marker type (Fig. 12A), the species the marker was developed in, or the species the marker was mapped in. To find out more information about the marker types, click the question mark logo next to Marker Type. For markers that have been mapped to a genetic map, the search can be restricted to a certain linkage group (Fig. 12B). And the search can be even further restricted to a certain locations on the linkage group.

Figure 12. CGD Marker Search interface
The search results are returned in a table format (Fig. 13). There are hyperlinks within the table that take you to more details about the marker or where the marker is mapped. This data table can be downloaded in a format that can be easily opened in applications such as Excel (Fig. 13A). Clicking on the marker name (Fig. 13B), displays the marker page which has information such as alignments and map positions. If you would like to change the search, either edit the parameters or click the reset button to start all over.
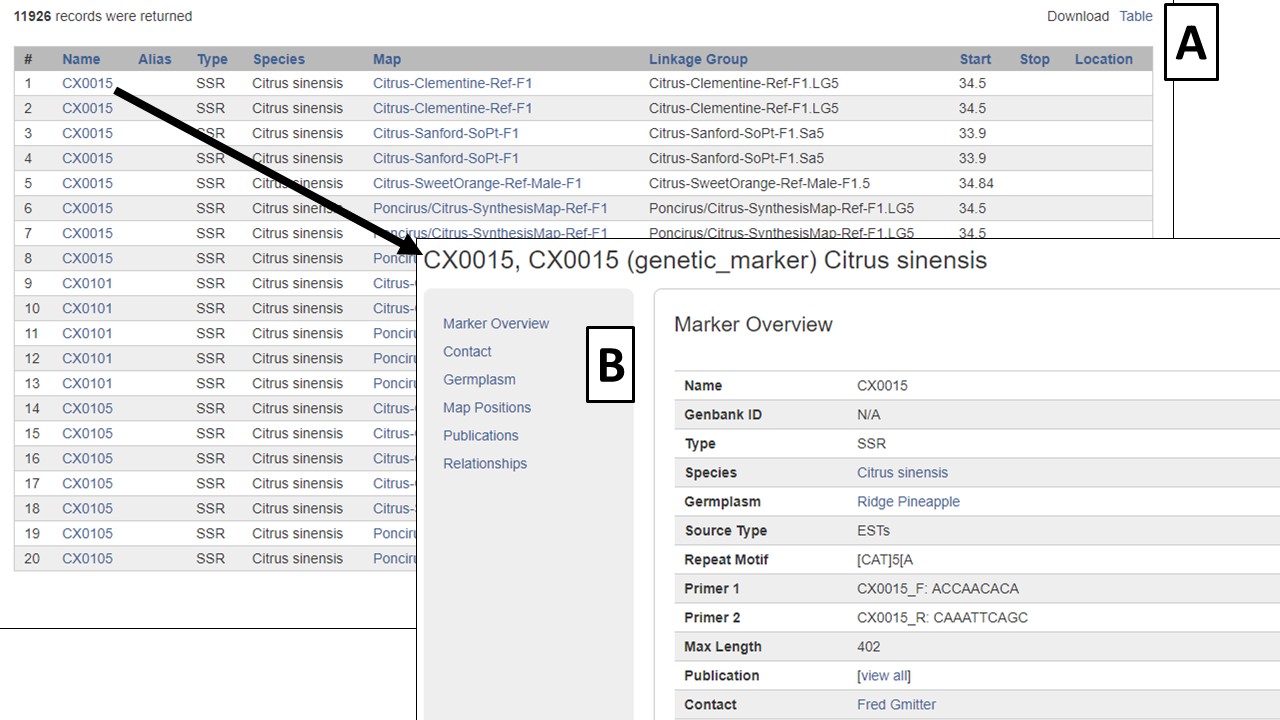
Figure 13. Marker Search results
Publications
To search publications on CGD, click on the Search menu and select Publication Search. The Publication Search is like many common literature searches, but is limited to publications that have been added to CGD. The publications can be searched by using keywords within different fields (Fig. 14A). Multiple fields and keywords can be entered and more fields can be added by clicking the Add/Remove buttons (Fig. 14B). A range of years can also be entered (Fig. 14C).
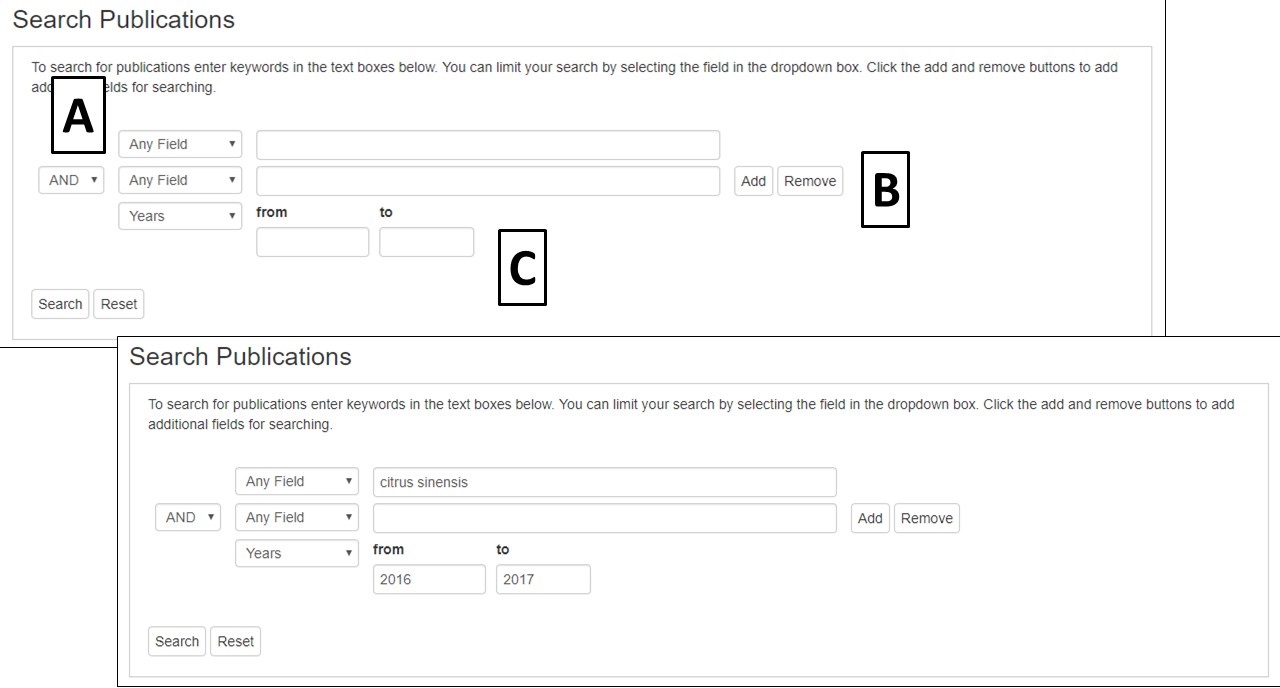
Figure 14. CGD Publication Search interface
The search results table has information about each publication (Fig. 15A). By clicking the publication title, more detailed information is displayed. Most publications also have a link to the publisher website or PubMed record in their titles (Fig. 15B).

Figure 15. Publication search results and detailed publication information
QTLs and MTLs
Quantitative trait loci (QTLs) and Mendelian trait loci (MTLs) that are entered in CGD are searchable from the QTL Search option under the Search menu in the header. The search can be restricted by QTL or MTL as well as by species (Fig. 16A). A certain trait ontology category can be selected to limit the returned results to only that trait type. If you are looking for a specific trait, you can do a keyword search of the trait names or search by the published name or the CGD assigned name (Fig. 16B). Please see the CGD Trait Abbreviation Table for more information about the trait abbreviations used in CGD.

Figure 16. CGD QTL Search interface
The results are returned in a table that has blue hyperlinks to more information about the QTL or MTL, the map it is located on, and the species it is from (Fig. 17). The table can also be downloaded (Fig. 17A). Clicking on the QTL label, opens the QTL information page which has a link to map positions (Fig. 17B). From the map position table, the QTL can be viewed in MapViewer or CMap.

Figure 17. CGD QTL Search results
Sequences
To search sequences, click on the Search menu on the homepage header and select Sequence Search. This search is useful for retrieving information about certain sequences from a larger dataset, or all the sequences from one or more datasets. The Gene and Transcript Search page is for searching genes and transcripts only, whereas the Sequence Search page also includes other sequence types. A few search parameters can be used to return a broad range of results, or more parameters can be selected to find very specific data. This is not a BLAST search, BLAST is available under the Tools menu.
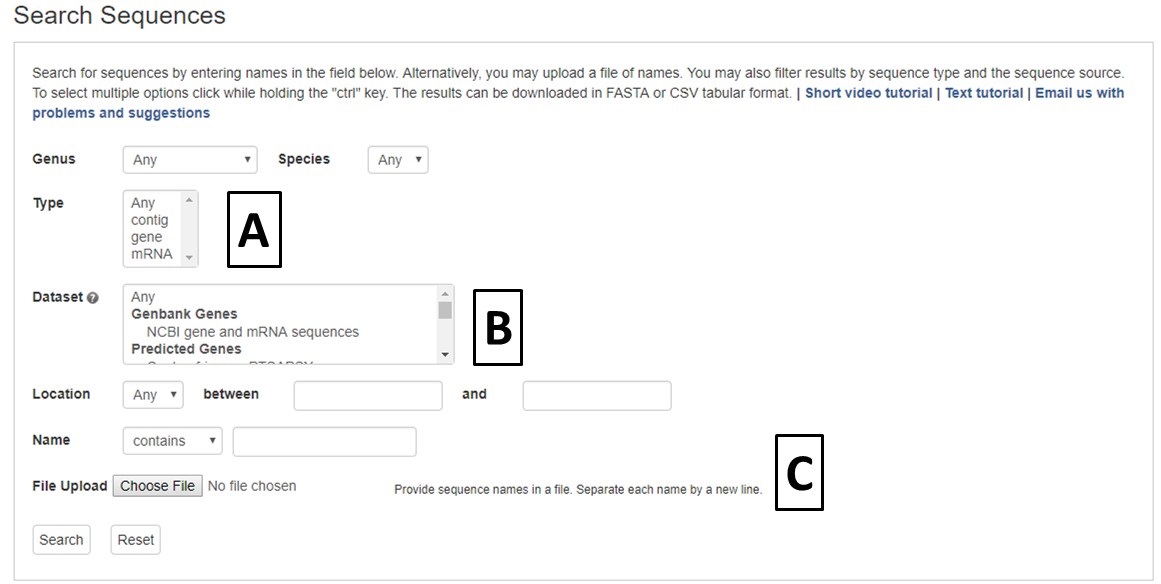
Figure 18. CGD Sequence Search interface
Sequence searches can be limited by genus and species, or sequence type (Fig. 18A). The search can also be restricted to a certain genome dataset and further restricted to a certain chromosome or scaffold location for within that assembly (Fig. 18B). The sequences in CGD can also be searched by sequence name, and there is an option to upload a text file of sequence names (Fig. 18C).

Figure 19. Search results table
All search results are returned in a table with hyperlinks to more information (Fig. 19A). The results table can be downloaded and a Fasta file of the sequences on the table can also be downloaded (Fig. 19B). To do a different search, either edit the parameters and search again, or click the Reset button.
Tools
To access the different Tools available on CGD, click on the Tools menu in the header and then select the tool you want to use (Fig. 20A). Many of the tools are also quickly accessed through links in the Tools Quick Start (Fig. 20B). To learn more about each search tool, please see the links below the figure.

Figure 20. Tools menu in header and Tools Quick Start on homepage
BLAST
CGD offers BLAST with sequence databases from sweet orange, clementine, mandarin, and trifoliate orange. Genome, unigene, and reference transcriptome assemblies for the citrus species are available along with complete genomes of Ca. Liberibacter and Liberibacter crescens. The BLAST interface looks and functions like the interface available on NCBI and information on the different settings and how to use BLAST can be found in the BLAST Help manual. The new Tripal BLAST module displays the results in a interactive interface (Fig. 21A) and for alignments to the genome scaffolds that are also in the CGD JBrowse, there is a link to view the BLAST hit in JBrowse (Fig. 21B). Features that are in the CGDdatabase (CDS, peptides, unigenes, RefTrans), will have links to more information in CGD.
.jpg)
Figure 21. BLAST results on CGD
JBrowse
CGD has an instance of the JBrowse genome browser for viewing genome data. A list of the genomes available in CGD can be accessed by clicking the JBrowse link in the Tools menu. Please watch the JBrowse tutorial for more details about how to navigate and use JBrowse.
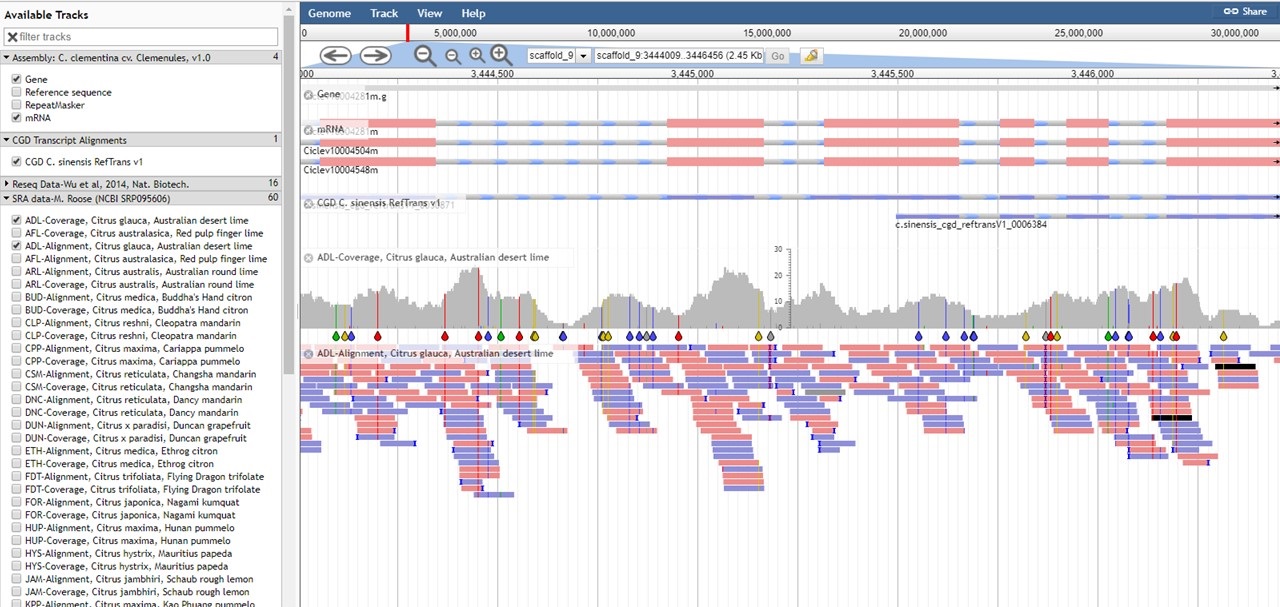
Figure 22. JBrowse of C. clementina genome.
For the tracks of aligned reads (BAM files) in the C. clementina v1.0 genome, the table below describes what the read colors mean in JBrowse.
| Aligned Read Color Meaning | Color of Read |
|---|---|
| Forward Strand | |
| Reverse Strand | |
| Forward strand missing mate | |
| Reverse strand missing mate | |
| Forward strand not proper | |
| Reverse strand not proper | |
| Forward strand on different chromosome | |
| Reverse strand on different chromosome |
CitrusCyc
CitrusCyc pathways were generated using Pathway Tools and are available under the CitrusCyc link in the Tools menu. Pathway Tools allows users to view metabolic pathways that are in genomes. Please see the manual for Pathway Tools for more information on use.

Figure 24. Citrus clementina cellular overview in CitrusCyc on CGD
MapViewer
MapViewer is a new tool for viewing genetic maps on CGD. It can be accessed from the Tools menu in the header (Fig. 25A), the Species Overview page (Fig. 25B), or the Map Overview page. The Map Overview page displays a summary graphic of all linkage groups (Fig. 25C) and clicking a linkage group opens a more detailed view in MapViewer.

Figure 25. Different ways to open MapViewer.
MapViewer displays the complete linkage group on the left, and the selected region on the right (Fig. 26). The selected region can be changed by dragging and resizing a window on the complete linkage group on the left side. There is a legend of the marker colors below the linkage group figure (Fig. 26A). Information about the markers is displayed in the upper right corner when the pointer is over a marker name on the right side graph. Clicking on the marker name on the rights side graph, opens the marker details page.

Figure 26. MapViewer displays a static linkage group graph on the left and a dynamic graph on the right.
A different map or linkage group can be displayed using the controls at the bottom of the MapViewer page (Fig. 27A). The color of the markers and which markers are displayed can be changed with the controls (Fig. 27B). The ruler and marker positions can also be toggled on or off (Fig. 27C). After changing any of the four parameter sections, the Submit button must be pressed to display the changes.
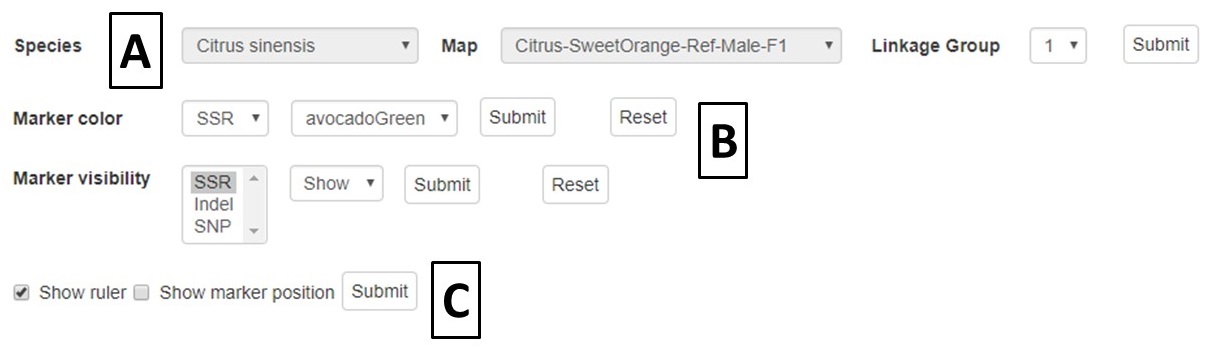
Figure 27. MapViewer control panel.
Pictures from MapViewer
MapViewer allows users to export figures as a high-resolution PNG file that is suitable for publication. Items that can exported have a clickable camera icon that will trigger a file download. Users can export the map overview which has all the linkage groups (Fig. 1), a single linkage group (Fig. 2), a linkage group comparison (Fig. 3), a dot plot (Fig. 4) or a correspondence matrix (Fig. 5).

Figure 1. How to download a map overview.

Figure 2. How to download a linkage group figure.
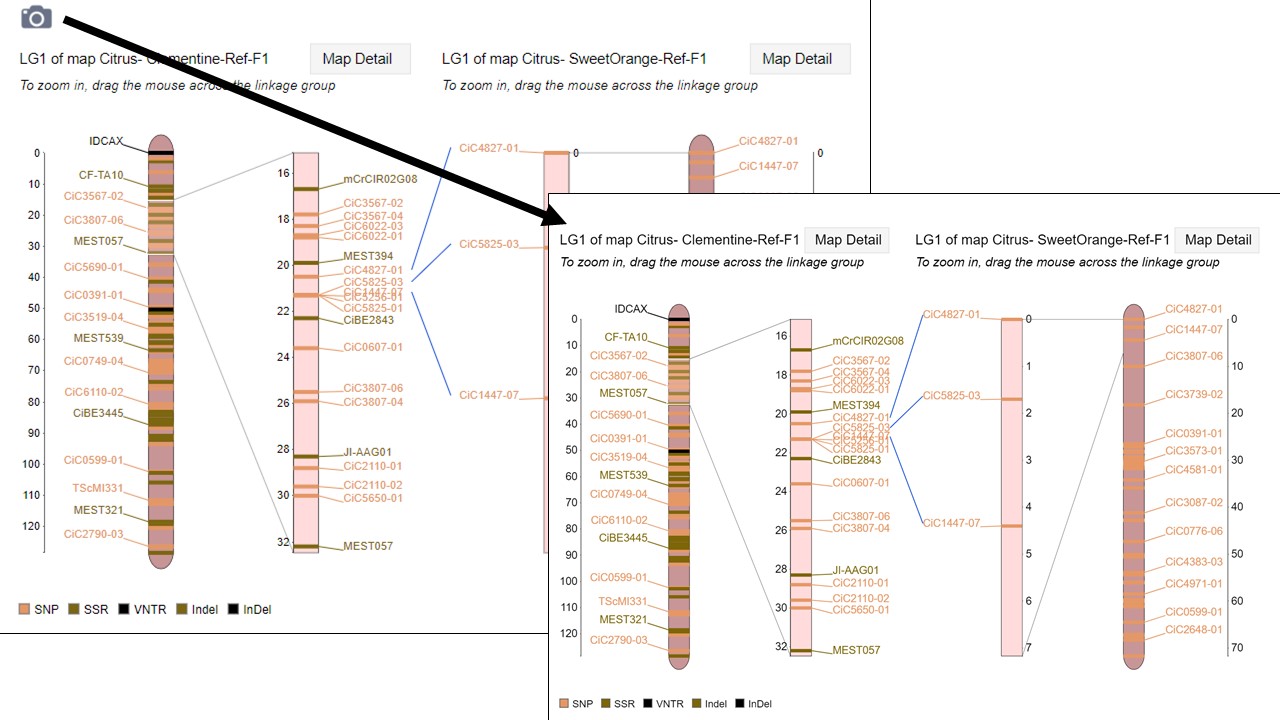
Figure 3. How to download a linkage group comparison figure.

Figure 4. How to download a linkage group comparison dot plot.

Figure 5. How to download a linkage group comparison correspondence matrix.
Synteny Viewer
CGD uses the Tripal Synteny Viewer, developed by the Fei Bioinformatics Lab, to display the analysis results of Citrus and Liberibacter genomes that were compared using the program MCScanX. Synteny Viewer is accessed under the "Tools" menu (Fig. 1A) or via a link on the Species Overview page.
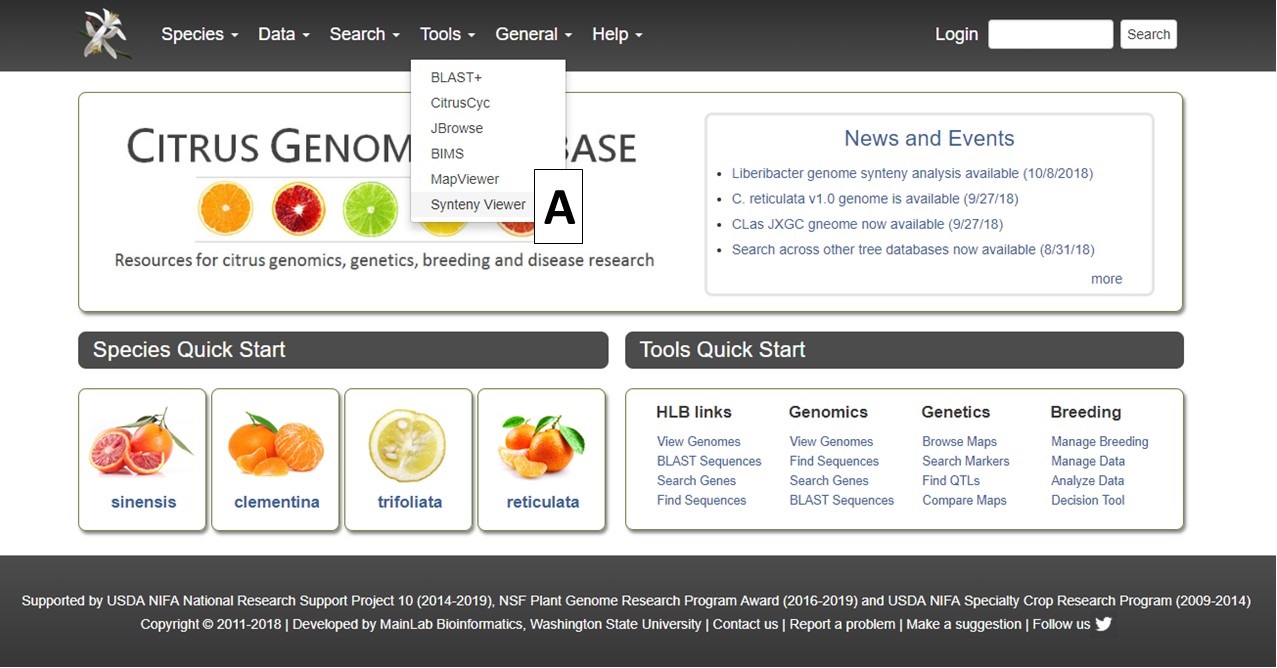
Figure 1. Accessing Synteny Viewer on CGD.
On the Synteny Viewer interface, there is some information about the tool and a pull-down menu to select the Organism Type (Fig. 2A) of "Plant" or "Bacteria" (Fig. 2B). The "Plant" option allows for the comparison of Citrus sp. genomes and the "Bacteria" option is for comparing genomes of the Ca. Liberibacter sp. and Liberibacter sp. If the block ID number is already known, the block ID value can be input directly to return just those results.

Figure 2. Selecting plant or bacteria genome option.
Once the organism is selected, another pull-down menu becomes available to select the first genome for comparison (Fig. 3A). After selecting the genome, the "Chromosome/Scaffold" menu will populate with the names of the appropriate scaffolds or chromosomes and one of the sequences can be selected (Fig. 3B). The final option is to select one or more genomes for comparison (Fig. 3C), and then the "Search button" is clicked to start the search (Fig. 3D).
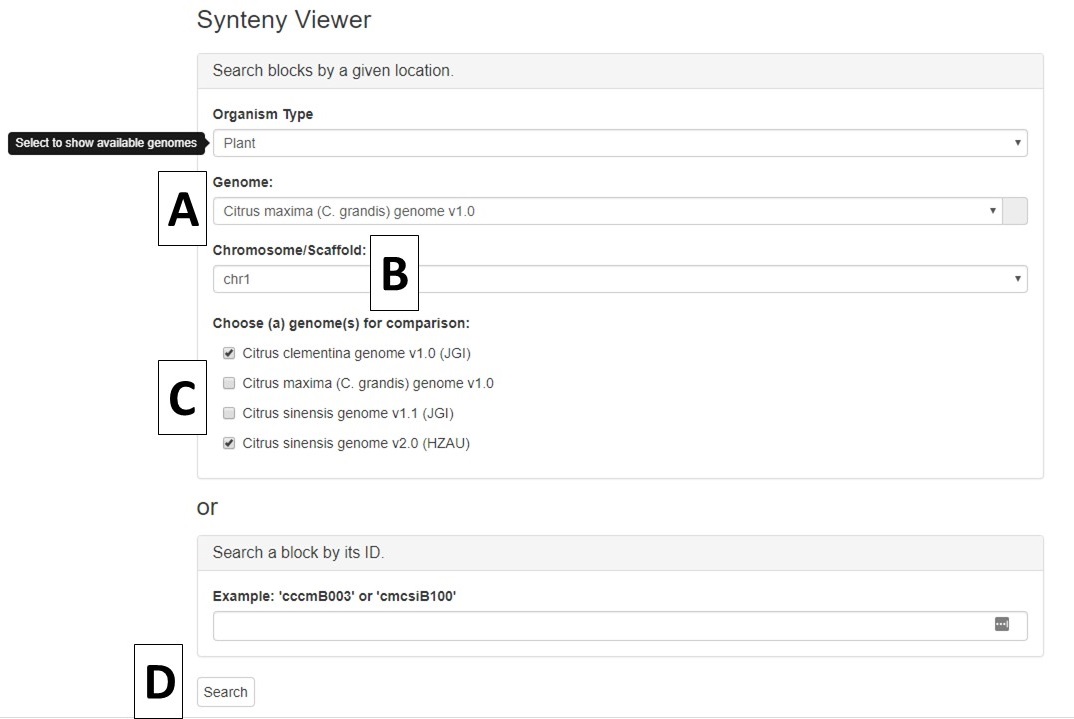
Figure 3. Options for Synteny Viewer search.
When the search is complete, a new page opens with the results. There is a summary of the input settings at the top of the page (Fig. 4A). When multiple genomes are queried, there are tabs to switch between the results (Fig. 4B) and the circular graph (Fig. 4C) will change when a different genome is selected. The syntenic regions are indicated on the circular graph by gray lines. When the mouse hovers over the gray line, a summary is displayed (Fig. 4D). Clicking on the gray lines, opens a page with more details (see below, Fig. 6).
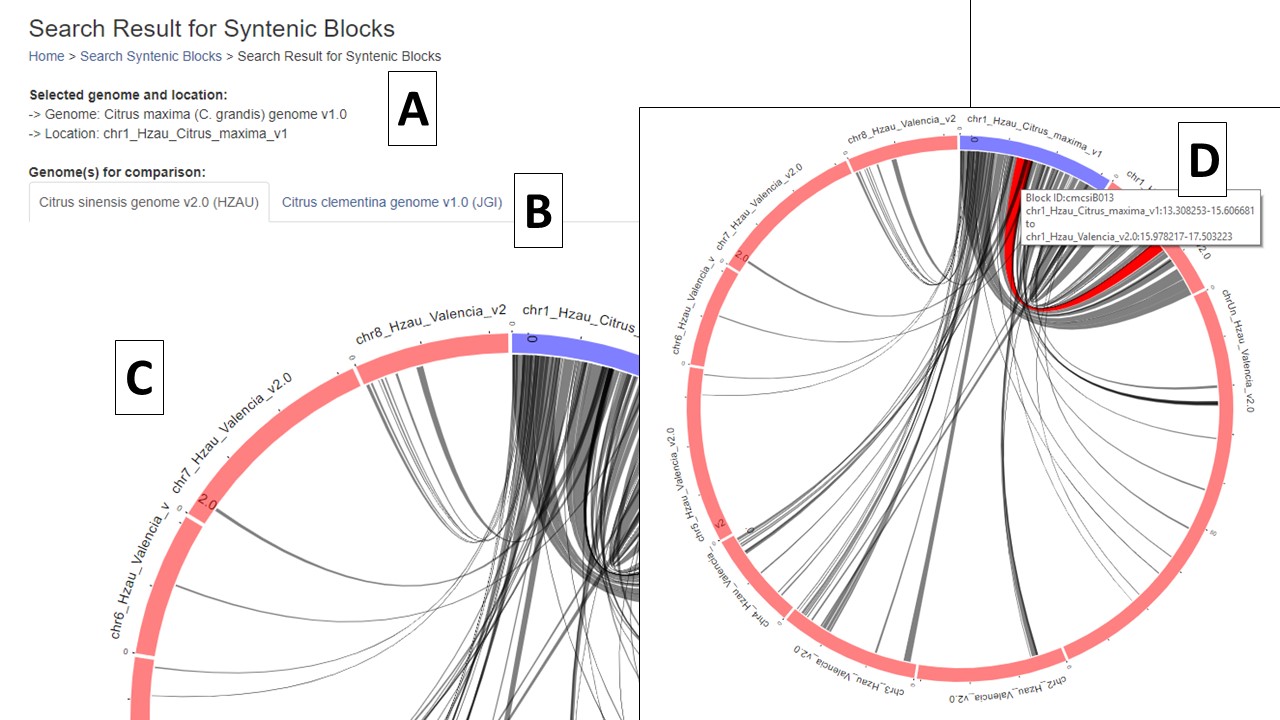
Figure 4. Synteny results and circular graphs.
Under the circular graph is a table listing the syntenic blocks that are displayed as gray lines in the circular graph. Clicking on the block name (Fig. 5A) opens the same details page as clicking on the gray lines on the circular graph.
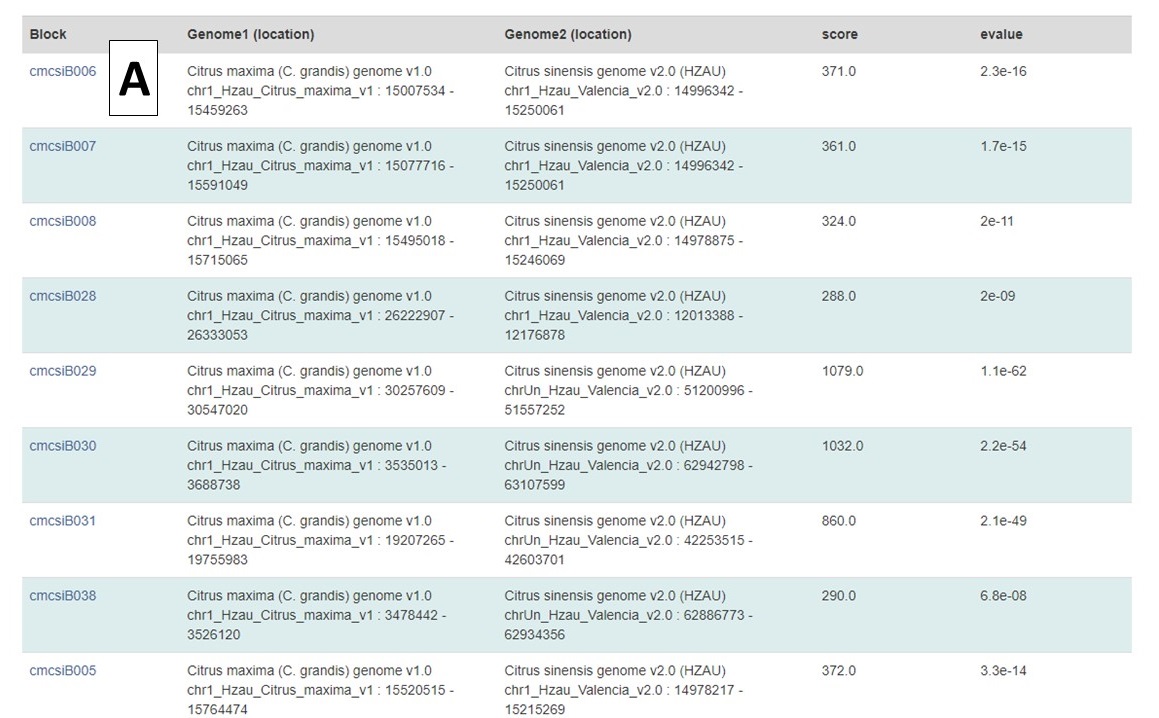
Figure 5. Synteny block list.
The syntenic block details page has an overview section at the top listing the details for each genome in the comparison (Fig. 6A), a side-by-side graphic of the syntenic block from each genome (Fig. 6B), and a table showing the genes in the syntenic block from each genome (Fig. 6C). Clicking on the gene name in either the table or on the graphic will open the feature details page from the CGD database.
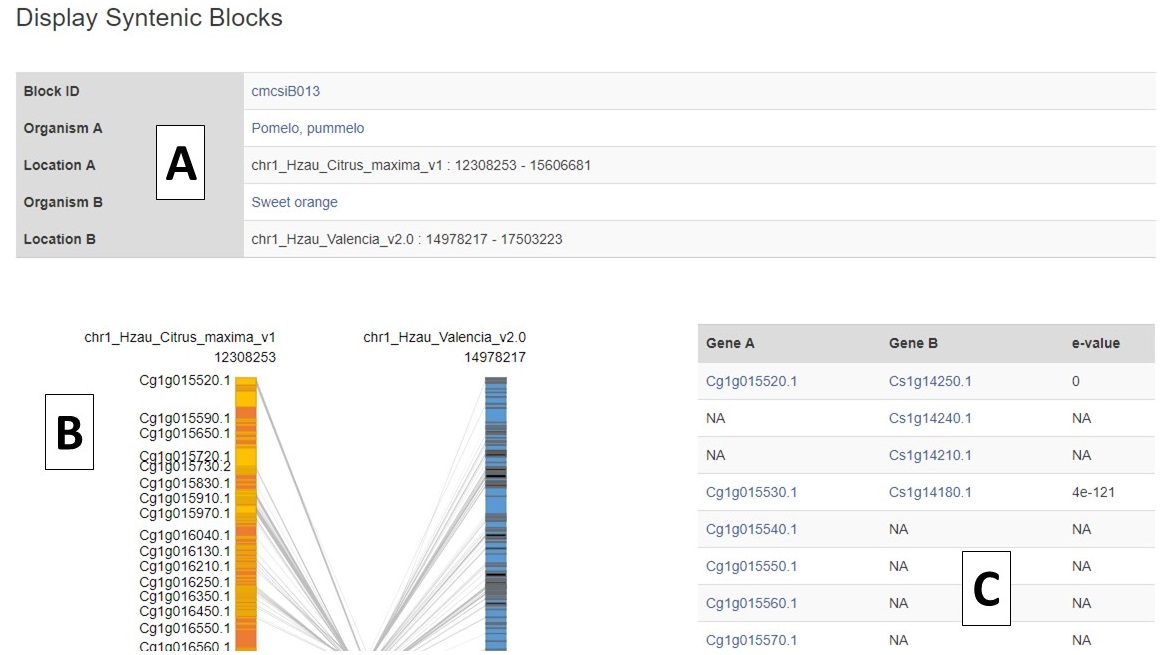
Figure 6. Syntenic Block Details Page.
The side-by-side graph can be zoomed (Fig. 7) using the scroll wheel on the mouse and the view can be shifted up or down by click and dragging.
.jpg)
Figure 7. Zoomed in view of graph.
CGD Video Tutorials
Please check out the tutorial videos for our other databases on the Main Lab YouTube channel. The searches and tools use the same framework across all of our databases.
-
Searches
-
Tools
-
Advanced queries and data retrieval
- How to download markers from map region
- How to view genomes linked to genetic maps
- How to go from BLAST to mRNA to JBrowse
- How to go from trait to GWAS to gene to ortholog
- How to find a marker associated with a trait
- How to download protein sequences
- How to view correspondences between genomes and genetic maps
Or you can watch the entire playlist below.